Study on Early Screening Methods and Prediction Models for
Colorectal Cancer Based on Biomarkers of Gut Microbiota
Background
Colorectal cancer (CRC) has the third highest cancer incidence in the word[1]. Early screening of CRC could sharply level up survival rate of patients according to numerous clinical practice, whereas current detection methods such as colonoscopy and fecal occult blood test (FIT) are far from satisfactory due to poor specificity and compliance[2]. Gut microbiota is believed to initially involved in colorectal carcinogenesis, indicating a potential role of microbial marker in early screening of CRC[3]. However, current discoveries of microbial markers and prediction models are based on metagenomic profiling, limiting the large cohort screening and validation[4].
Results
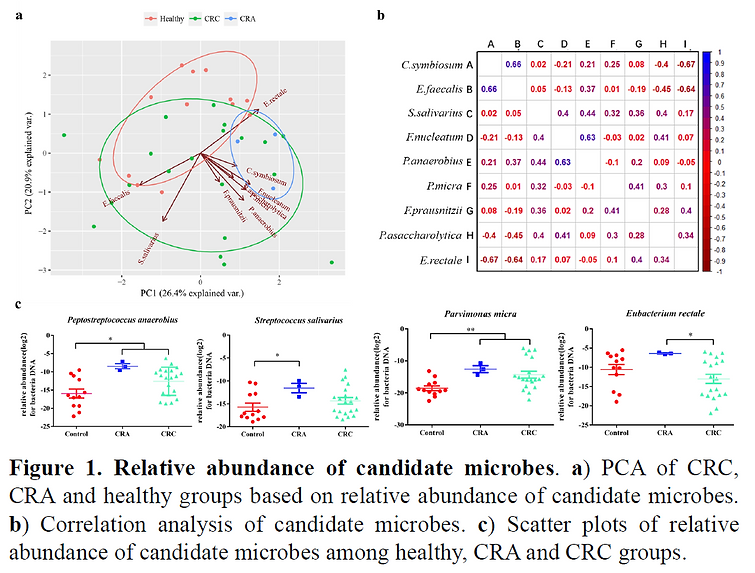
We found a separation among CRA, CRC and healthy groups based on relative abundance of nine candidate gut microbes, with core contributions from P.anaerobius, S.salivarius, P.micra and E.rectale (Fig.1a). Specifically, the relative abundance of P.anaerobius and P.micra showed an increase in CRA and CRC groups (p<0.05 and p<0.01 respectively), compared to healthy controls. Generally, the relative abundance of these four microbes were higher in CRA group than CRC group, suggesting a capacity of sensitively discerning CRA (Fig.1c). Besides, nine microbes showed positive correlation (C.symbiosum and E.faecalis, F.nucleaum and P.anaerobius) and negative correlation (C.symbiosum and E.rectale) (Fig.1b).
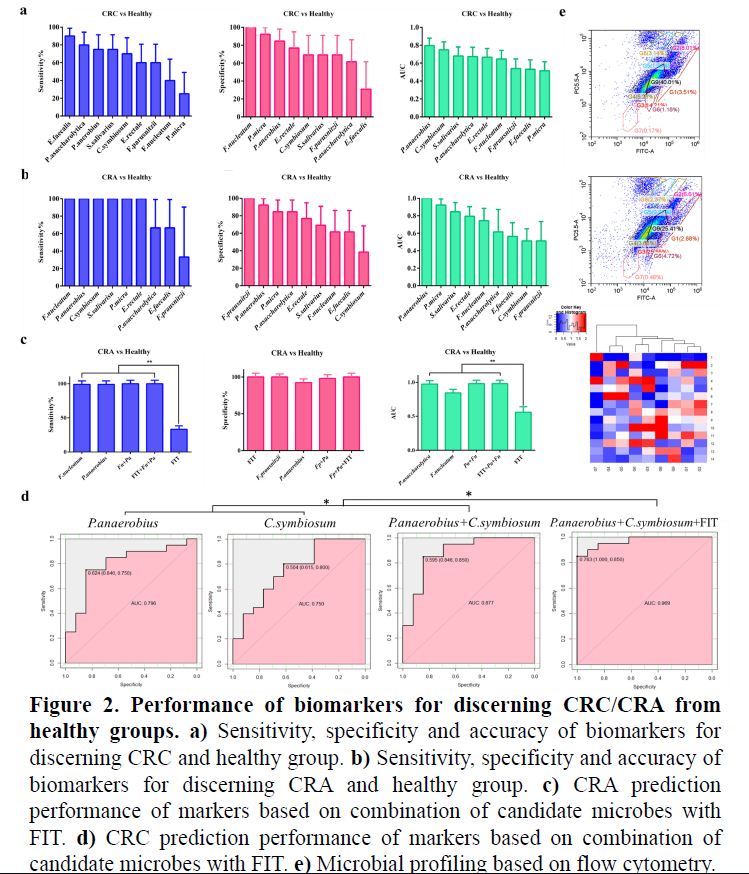
Biomarkers based on candidate gut microbes showed differently sensitivities and specificities when discerning CRC/CRA from healthy controls (Fig.2a,2b), among which, P.anaerobius and C.symbiosum exhibited best predictive accuracy. A combination of FIT and multiple candidate microbes could level up classification ability (Fig.2c,2d). Microbial clustering and similarity analysis based on flow cytometry data provide a possibility to analysis CRC-related microbial community (Fig.2e).
Methods
Text and data mining were conducted to score out nine candidate gut microbes from published articles and databases. A total of 46 fecal samples from voluntary CRC patients (n=19), colorectal adenoma (CRA) patients (n=3) and healthy volunteers (n=24) aging over 50 years old were recruited for fecal DNA extraction and qPCR amplification to detect the relative abundance of candidate gut microbes. PCA was applied to investigate candidate microbes’ difference at community level among healthy, CRA and CRC groups. Machine learning methods including Binary and multinomial logistic regressions, Naïve Bayes, SVM and random forest were applied to build prediction models for discerning CRA/CRC groups from healthy groups. Assessment of each model was based on ROC analysis.
Significance
• This study firstly demonstrated the potential of text-mined and experiment-validated gut microbes to predicting the risk of early colorectal cancer as well as adenoma.
• This study systematically compared and established predictive models for CRC and CRA based on machine-learning methods and data from multiple sources.
• This study firstly explored the correlation and co-occurrence among candidate microbes, setting the reference for further integration of multiple microbial markers.
Reference
[1]IARC. Cancer fact sheets: colorectal cancer [M].2018.
[2]QUINTERO E, CASTELLS A, BUJANDA L, et al. Colonoscopy versus Fecal Immunochemical Testing in Colorectal-Cancer Screening [J]. New England Journal of Medicine, 2012, 366 (8): 697-706.
[3]SEARS CYNTHIA L, GARRETT WENDY S. Microbes, Microbiota, and Colon Cancer [J]. Cell Host & Microbe, 2014, 15(3): 317-28.
[4]WANG J, JIA H. Metagenome-wide association studies: fine-mining the microbiome [J]. Nature Reviews Microbiology, 2016, 14:508-522.